Search option
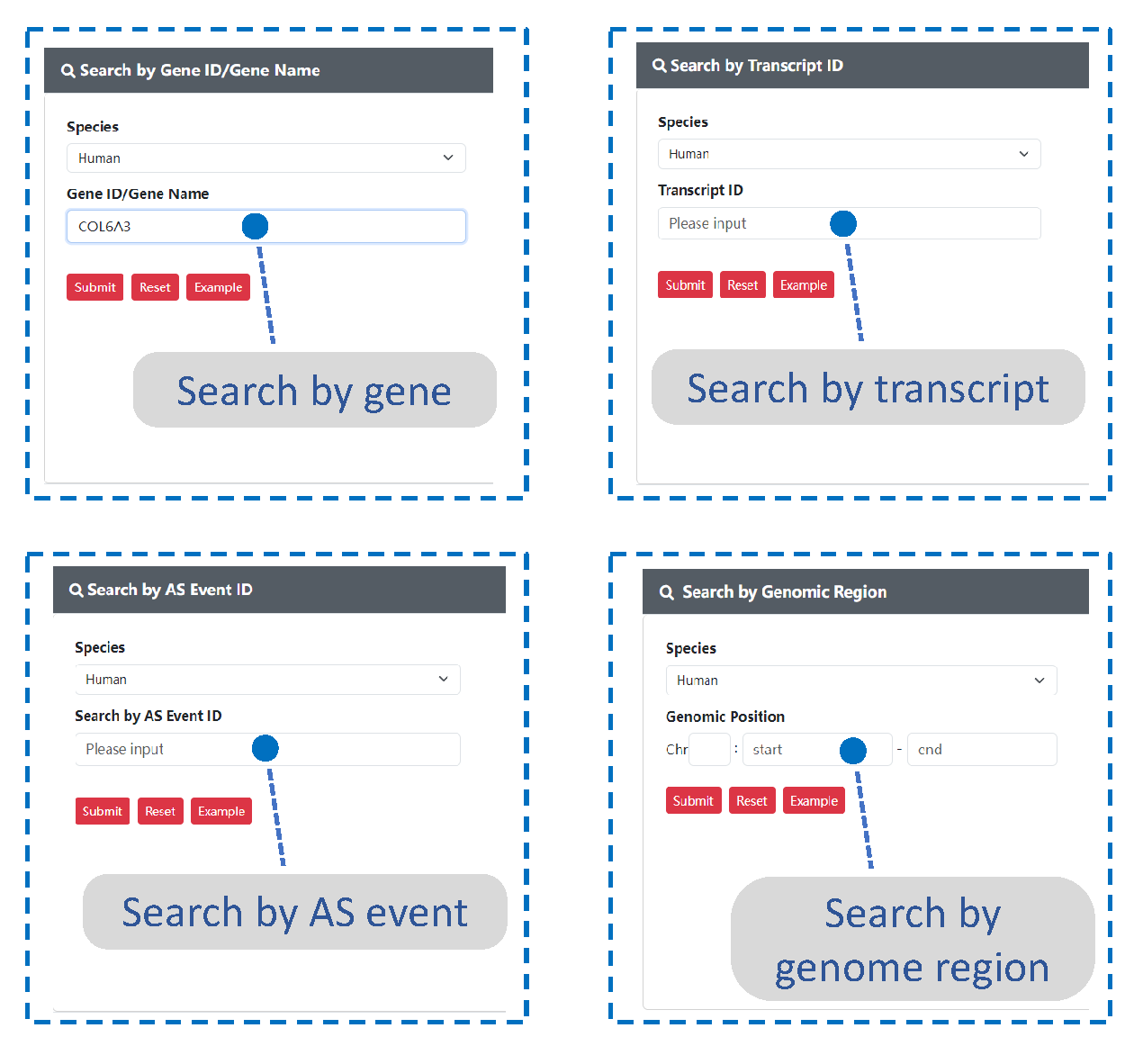
Table information
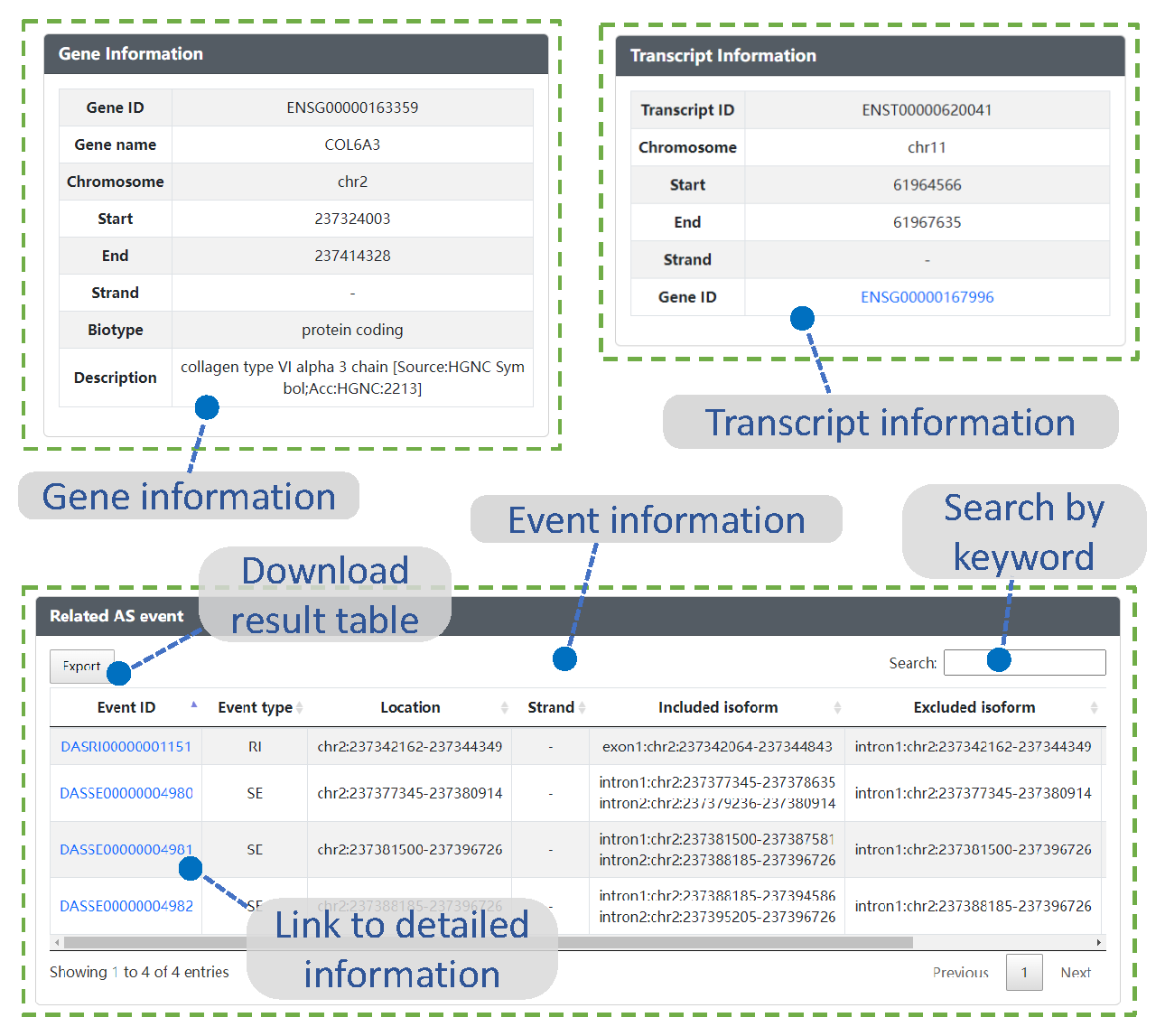
- Gene ID : Reference annotation gene ID, commonly Ensembl. The novel lncRNAs were named beginning with "DASES".
- Gene Symbol : HGNC gene symbol, if available, otherwise "NULL" or blank.
- Transcript ID : Reference annotation transcript ID, commonly Ensembl. The novel lncRNAs were named beginning with "DASES".
- Event ID : Alternative splicing event ID, generated by DASES.
- Event type : The type of alternative splicing event. Can be exon skipping (ES), intron retention (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), mutually exclusive exons (MXE).
- Chromosome : Reference genome chromosome name.
- Start : First position of gene, transcript, or alternative splicing event (1-based).
- End : Last position of gene, transcript, or alternative splicing event (1-based).
- Location : Chromosome name with first and last position of the alternative splicing event.
- Strand : Given as + or -.
- Biotype : Gene biotype.
- Description : Description of gene.
- Included isoform : Isoform including exon under the comparison of the two transcripts, specifically the shorter total length of the transcript intron as the included isoform Column.
- Excluded isoform : Isoform excluding exon under the comparison of the two transcripts, specifically the longer total length of the transcript intron as the excluded isoform Column.
- Overlapped protein domain : Protein domains that intersect with the alternative splicing event at genomic locations.
- Related transcript ID : Transcript related to the alternative splicing event.
Figure information
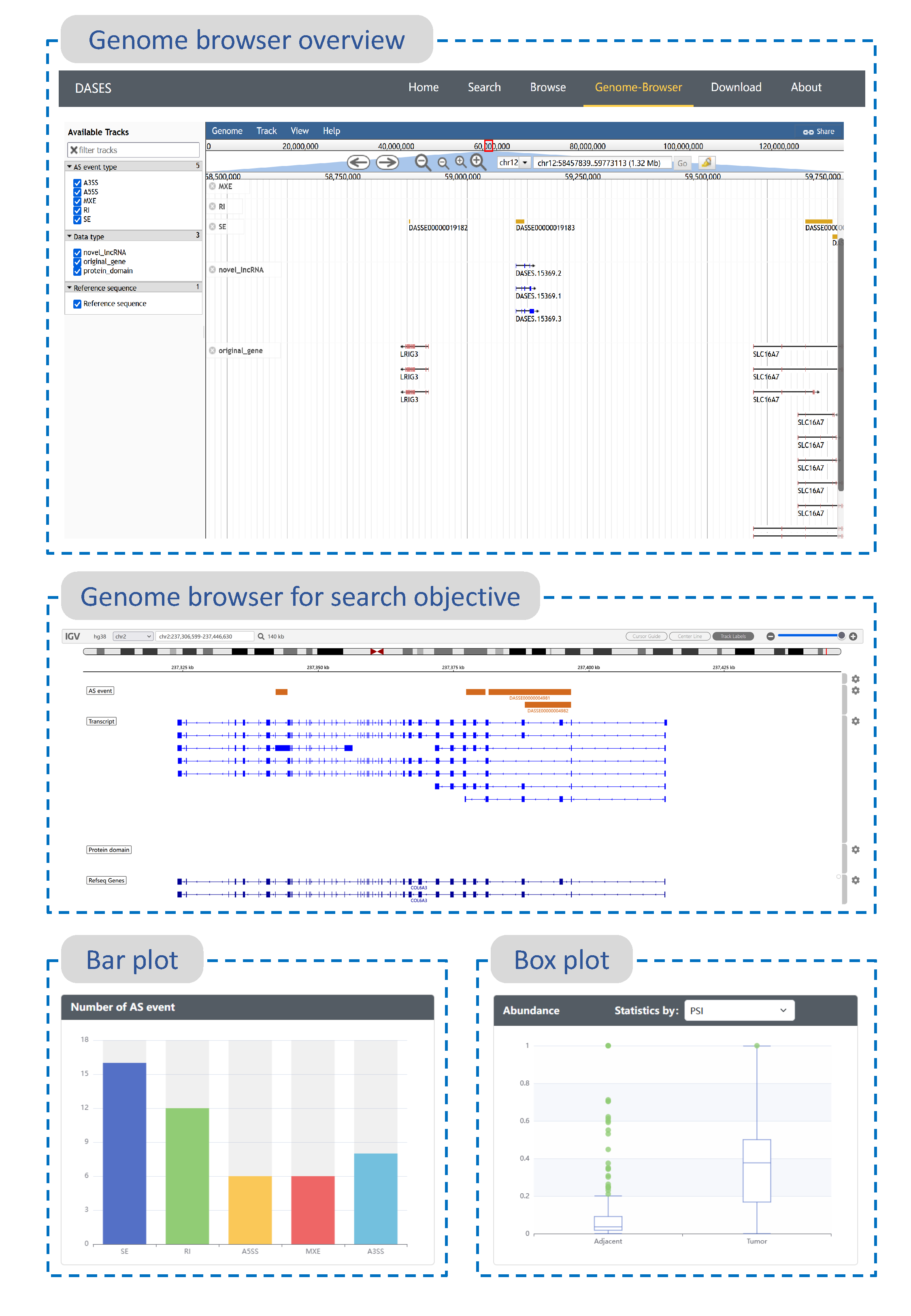
- Genome browser overview : The genome browser provides a visual representation of all identified alternative splicing events and their associated genes, transcripts, and protein domains. Built on the JBrowse software, the browser offers five different tracks for different types of alternative splicing events. Additionally, tracks for genes, transcripts, and protein domains are included, each assigned a distinct color for easy differentiation. Users can explore specific regions of interest using this browser.
- Genome browser for search objective : This genome browser, created using the IGV software, is designed to display the search results for specific search objectives such as genes, transcripts, or alternative splicing events. Users can input their search queries through the search interface, and the browser will present the searched objects along with their associated genes, transcripts, and alternative splicing events in result page.
- Bar plot :This bar plot visually represents the statistical distribution of different types of alternative splicing events associated with a specific search objective, which can be a gene or a transcript.
- Box plot : This box plot illustrates the statistical distribution of Percent Spliced In (PSI), Inclusion Junction Counts (IJC), and Exclusion Junction Counts (EJC) values for the specific alternative splicing events of interest across all tumor and adjacent normal tissue samples.